项目文章
2014-Neurpscience-IF3.33-Proteomics reveals energy and glutathione metabolic dysregulation in the prefrontal cortex of a rat model of depression

Abstract
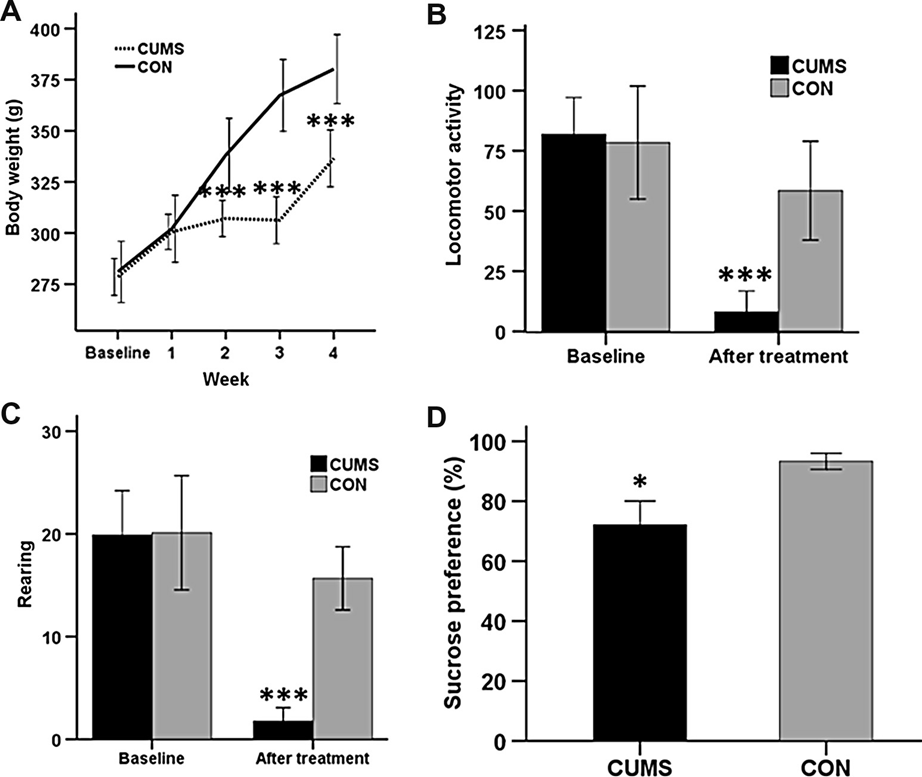
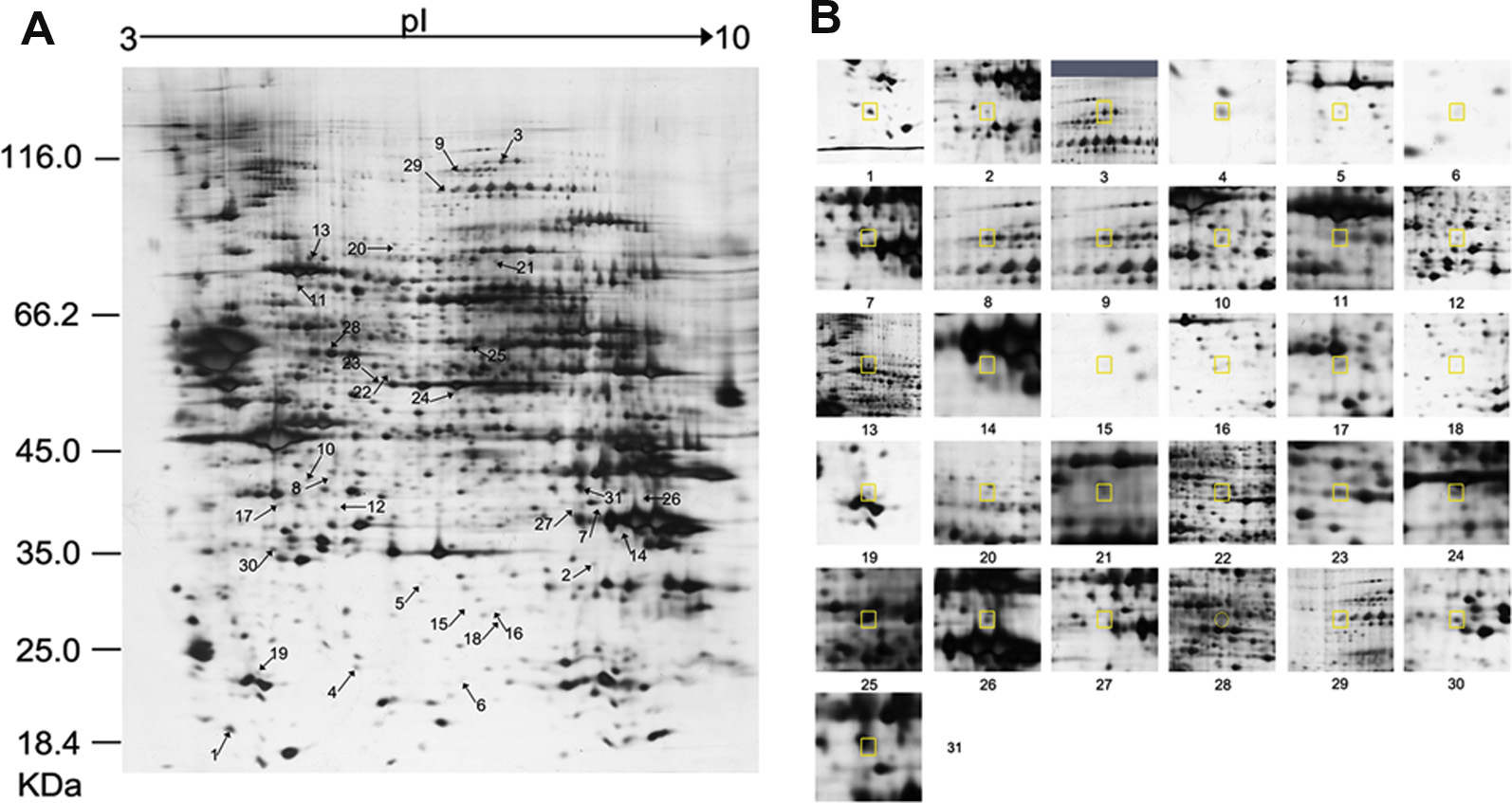
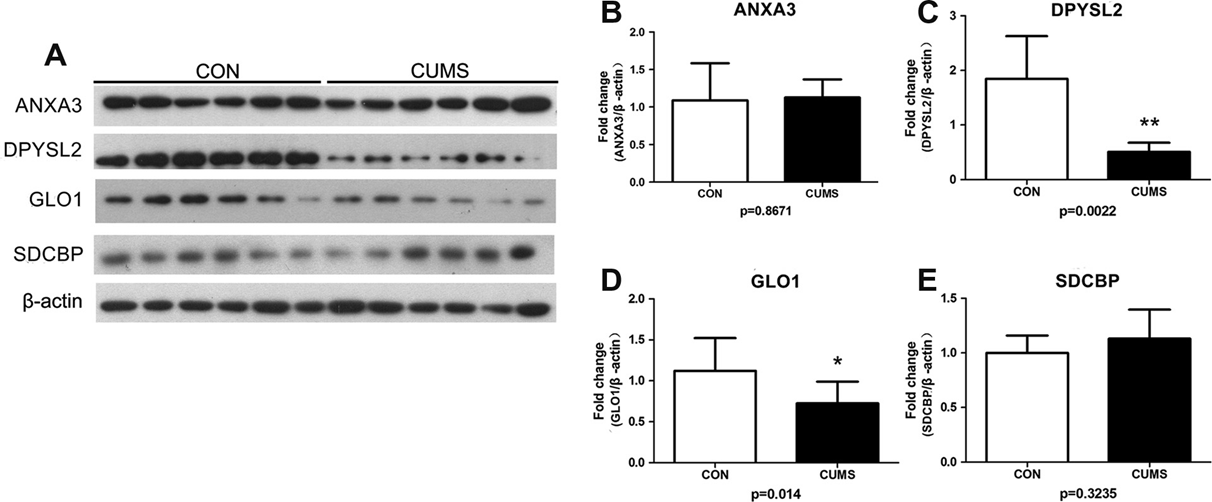
Major depressive disorder (MDD) is a prevalent debilitating psychiatric mood that contributes to increased rates of disability and suicide. However, the pathophysiology underlying MDD remains poorly understood. A growing number of studies have associated dysfunction of the prefrontal cortex (PFC) with depression, but no proteomic study has been conducted to assess PFC protein expression in a preclinical model of depression. Using the chronic unpredictable mild stress (CUMS) rat model of depression, differential protein expression between the PFC proteomes of CUMS and control rats was assessed through two-dimensional electrophoresis followed by matrix-assisted laser desorption ionization-time of flighttandem mass spectrometry. Differential protein expression was analyzed for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway over-representation. Four differential proteins were selected for Western blotting validation.Twenty-nine differential proteins were identified in the PFC of CUMS rats relative to control rats. Through KEGG analysis, energy and glutathione metabolic pathways were determined to be the most significantly altered biological pathways. Two of the four differential proteins selected for Western blotting validation – glyoxalase 1 and dihydropyrimidinase- related protein 2 – were found to be significantly downregulated in CUMS relative to control rats. In conclusion,proteomic analysis reveals that energy and glutathione metabolism are the most significantly altered biological pathways in the CUMS rat model of depression. Further investigation on these processes and proteins in the PFC is key to a better understanding of the underlying pathophysiology of MDD. Crown Copyright 2013 Published by Elsevier Ltd. All rights reserved.
文章链接:

